 |
 |
|
 |
 |
 |
QUILTS
is a tools for creating sample specific protein sequence databased. It uses genomic and transcriptomic information to create comprehensive sample specific protein database that supports the identification of novel proteins, resulting from single nucleotide variants, splice variants and fusion genes.
|
|
GPMDB
is a database of tandem mass spectra and their assigned peptide sequences.
It is designed to aid in the difficult process of validating peptide MS/MS spectra.
|
 |
 |
 |
 |
OpenSlice is a novel web-based API for large scale data sharing in Mass Spectrometry fully eliminating the need to download RAW datafiles.
|
|
Llama Magic is a tool for identification of single chain Llama antibodies by combining next generation sequencing data with mass spectrometry based affinity proteomics data.
|
 |
 |
 |
 |
PGx is a tool for proteogenomics mapping. It takes peptide identities and quantities and maps them onto the human genome.
|
|
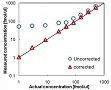
SRM Interference is a tool for detecting and correcting for interference in SRM analysis, and for detecting the linear range of calibration curves.
|
 |
 |
 |
 |
Copurification supports the curation of images displaying protein co-purification banding patterns resulting from affinity capture followed by e.g. SDS-PAGE and protein staining. The conditions of each affinity capture experiment curated here are linked to the resulting banding pattern so that purifications carried out in different conditions and/or by different researchers can be effectively searched and compared to one another.
|
|
FLIP is a novel assay for fast and reliable evaluation of an immunoprecipitation reaction that relies on the expression of the target protein as a chromophore-tagged protein and couples IP with the measurement of fluorescent signal coating agarose beads.
|
 |
 |
|
 |
 |

